Open Source Bioreactor Project
We are building an open source, benchtop, batch bioreactor to optimise yield of enzymes producing recombinant proteins for molecular biology such as Taq polymerase or for cell-free extract production. This will build on existing open source projects to further reduce the cost of components and pay particular attention to their global accessibility. By generating a modular design with thorough and useful documentation of different options to suit budget and accuracy requirements, we will make these devices easier to build and maintain for a wider range of users in universities, companies and biomaker spaces: in particular those in resource-constrained contexts.
Contact us via Jenny Molloy (

Project Resources
Communications
- Chat Channel
- Meeting Room – next meeting TBC
- Project page at Biomakespace website
- Project page at biomaker.org: TBC
Documentation
Background
Bioreactor Team
Module Documentation




You can directly jump to the sections:
- The Problem
- Biological System
- Hardware Design Goals
- Project Implementation
- Outcome and Benefits
- References
- The TEAM
The Problem
The problem to be addressed is reliable culturing of bacteria for expressing lab-scale quantities of recombinant bacteria or for making cell-free extracts. Such cultures are known to vary in growth rate and other parameters which affect final yield or viability, so continuous monitoring and optimisation of growth conditions can improve reproducibility and reliability of experiments. Existing commercial solutions are prohibitively expensive for low-resourced labs and do not allow for easy customisation and maintenance. Existing open source designs typically fall into two categories:
a) small scale systems for parallel evolution experiments or characterisation of gene expression for synthetic biology (e.g. theKlavin’s Lab turbidostat and Aachen iGEM 2015 designs).
b) large scale systems such as the 8 L Aeronaut Brewing Company Bioreactor and 12 L home-built fermenter described by Uwe et al. (2008).
The EPFL/Univalle 1 L open source bioreactor is the most suitable starting point but there are several features that the current version is lacking to meet the design goals for our project and at an estimated cost of $1800, there is a need to more than halve the cost of the device to reach a budget of under £500 for the most feature-rich version of our bioreactor.
Biological Systems
We will use the workhorse of molecular biology Escherichia coli bacteria. In this project it will be expressing an off-patent Taq polymerase enzyme in an off-patent pTTQ18 vector that is readily available from Addgene (https://www.addgene.org/25712/). The goal is that the bioreactor is suitable for E. coli expressing other recombinant proteins and for other bacterial and yeast strains with parameter optimisation.
Hardware Design Goals
Sterile and autoclavable parts.
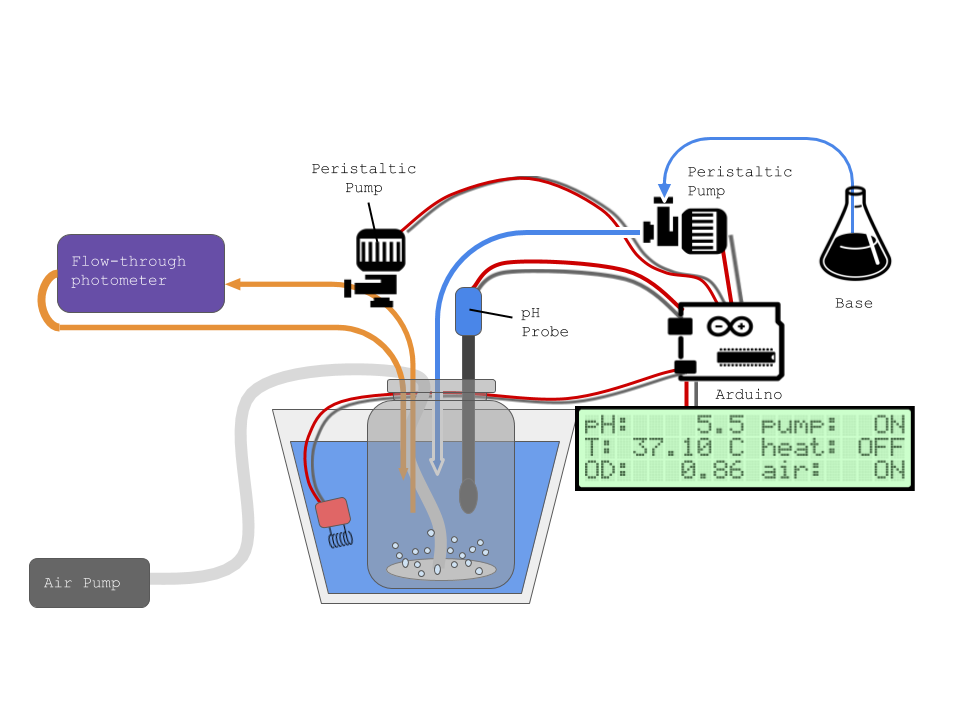
Control of temperature, pH, agitation and media supply. Measurement of weight and optical density.
Modular structure and use of parametric CAD designs to allow greatest flexibility for the user.
Open formats and licensing: All designs will be shared in open file formats that can be edited with free software and under maximally permissive open hardware/software licenses such as Solderpad/CERN OHL and MIT/Apache.
Useful-source: Documentation will be modular and not only sufficient to build the device but also to understand the functionality and design goals of each module. Where appropriate, we will present some possible alternative components to reduce cost or increase functionality or accuracy.
Easily sourced: we will use commonly and globally available components wherever possible and seek to avoid vendor lock in.
Under £500 for most advanced version of this design (suggestions will be made for more expensive alternatives and their respective benefits).
Project Implementation
The project will be split into different modules to be prototyped, tested and documented:
A: Culture vessel: a glass, autoclavable bottle with connectors for the inputs and sensors. One design challenge is to source lower-cost connectors that maintain sterility.
B: Agitation System: Three main options to review to modify the existing base design, a magnetic stirrer, stirring paddle or bubble-based aeration using a aquarium pump (Yasumitsu et al., 2013).
C: Temperature control system: Requires a temperature sensor and heating pad in closed loop control plus measurement of temperature readings throughout the culture volume.
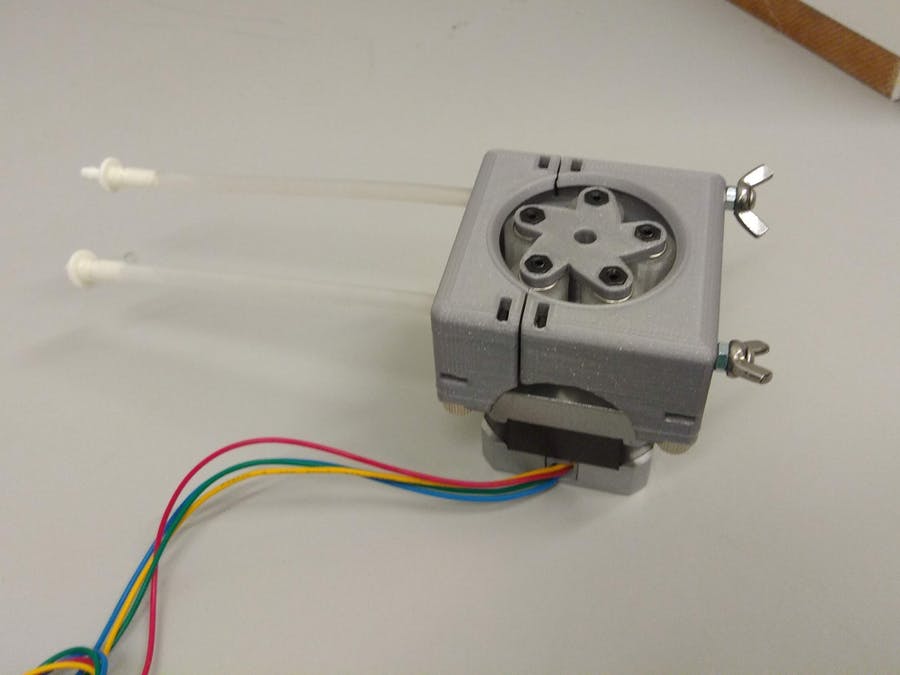
D: pH control system: We will test an online colorimetric sensor using immobilised neutral red (Hashemi et al., 2007) and a syringe pump driver or peristaltic pump to automatically adjust using NaOH.
E: Optical density sensor: The goal is to have a continuous monitoring device, there are several publications to review and test a range of sensors loops, taking the Aaachen 2015 iGEM team design as a starting point,
F: Controller system: The device will use an Arduino Mega or similar prototyping board to read sensors and control the pumps/heaters. Information will be transmitted via wifi for viewing in a browser, a small LCD screen will provide basic feedback and measurements to the user to allow monitoring.
The team will work in parallel on the different modules and have skills including biology, hardware and software. We will seek additional support from other teams and contacts where necessary, particularly for electronics.
Outcomes and Benefits
The outcomes of the project would be:
A physical bioreactor unit available at the Cambridge Biomakespace for anyone to use (for a small monthly fee), including future Biomaker Challenge teams.
Project report on Hackster with a fully documented build manual and a user manual for calibration and maintenance.
Open hardware designs shared on Docubricks and Github, open software on Github or other repository.
Increased knowledge and experience within the interdisiplinary team about specific technical areas e.g. electronic control of closed loop systems, plus broader ideas e.g. frugal innovation.
The benefits will be increasing capacity in low-resourced contexts for reliably growing microorganisms for local production of enzymes or other proteins and for producing cell-free extracts. This is important i) to increase access to research reagents at lower cost and without reliance on supply chains which may have challenges including shipping times and cold chain; ii) given the growing importance of cell-free extracts for research and education, low-cost diagnostics and more but the documented need to harvest bacteria at an appropriate growth stage to produce extract.
These benefits will be immediate for local researchers and community biology participants in terms of access to the equipment for a range of projects. The designs will be beneficial for those wanting to build their own bioreactor and will be shared internationally via the Gathering for Open Science Hardware community (400 members) and the Global Community Biosummit community (300 members) plus on Hackster.
References
- Riek, Uwe, et al. “An automated home-built low-cost fermenter suitable for large-scale bacterial expression of proteins in Escherichia coli.” BioTechniques 45.2 (2008): 187-189.
- Yasumitsu, Hidetaro, et al. “Fine Bubble Mixing (FBM) Culture of E. coli: A Highly Cost-effective Middle Scale-size Culture System.” Protein and peptide letters 20.2 (2013): 213-217.
- Hashemi, Payman, et al. “Agarose film coated glass slides for preparation of pH optical sensors.” Sensors and Actuators B: Chemical 121.2 (2007): 396-400.